R for reproducible scientific analysis
Split-apply-combine
Learning Objectives
- To be able to use the split-apply-combine strategy for data analysis
Previously we looked at how you can use functions to simplify your code. We defined the calcGDP function, which takes the gapminder dataset, and multiplies the population and GDP per capita column. We also defined additional arguments so we could filter by year and country:
# Takes a dataset and multiplies the population column
# with the GDP per capita column.
calcGDP <- function(dat, year=NULL, country=NULL) {
if(!is.null(year)) {
dat <- dat[dat$year %in% year, ]
}
if (!is.null(country)) {
dat <- dat[dat$country %in% country,]
}
gdp <- dat$pop * dat$gdpPercap
new <- cbind(dat, gdp=gdp)
return(new)
}A common task you’ll encounter when working with data, is that you’ll want to run calculations on different groups within the data. In the above, we were simply calculating the GDP by multiplying two columns together. But what if we wanted to calculated the mean GDP per continent?
We could run calcGPD and then take the mean of each continent:
withGDP <- calcGDP(gapminder)
mean(withGDP[withGDP$continent == "Africa", "gdp"])[1] 20904782844
mean(withGDP[withGDP$continent == "Americas", "gdp"])[1] 379262350210
mean(withGDP[withGDP$continent == "Asia", "gdp"])[1] 227233738153
But this isn’t very nice. Yes, by using a function, you have reduced a substantial amount of repetition. That is nice. But there is still repetition. Repeating yourself will cost you time, both now and later, and potentially introduce some nasty bugs.
We could write a new function that is flexible like calcGDP, but this also takes a substantial amount of effort and testing to get right.
The abstract problem we’re encountering here is know as “split-apply-combine”:
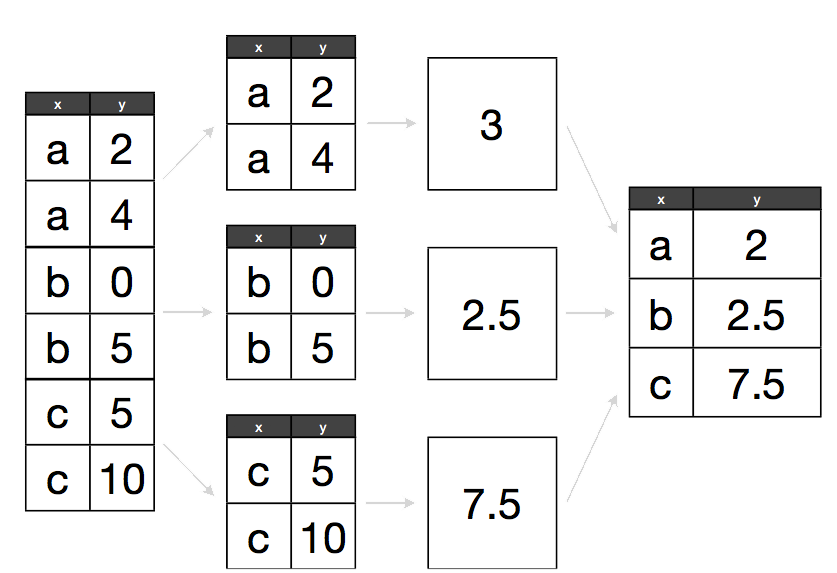
Split apply combine
We want to split our data into groups, in this case continents, apply some calculations on that group, then optionally combine the results together afterwards.
The plyr package
For those of you who have used R before, you might be familiar with the apply family of functions. While R’s built in functions do work, we’re going to introduce you to another method for solving the “split-apply-combine” problem. The plyr package provides a set of functions that we find more user friendly for solving this problem.
We installed this package in an earlier challenge. Let’s load it now:
library(plyr)Plyr has functions for operating on lists, data.frames and arrays (matrices, or n-dimensional vectors). Each function performs:
- A splitting operation
- Apply a function on each split in turn.
- Recombine output data as a single data object.
The functions are named based on the data structure they expect as input, and the data structure you want returned as output: [a]rray, [l]ist, or [d]ata.frame. The first letter corresponds to the input data structure, the second letter to the output data structure, and then the rest of the function is named “ply”.
This gives us 9 core functions **ply. There are an additional three functions which will only perform the split and apply steps, and not any combine step. They’re named by their input data type and represent null output by a _ (see table)
Note here that plyr’s use of “array” is different to R’s, an array in ply can include a vector or matrix.
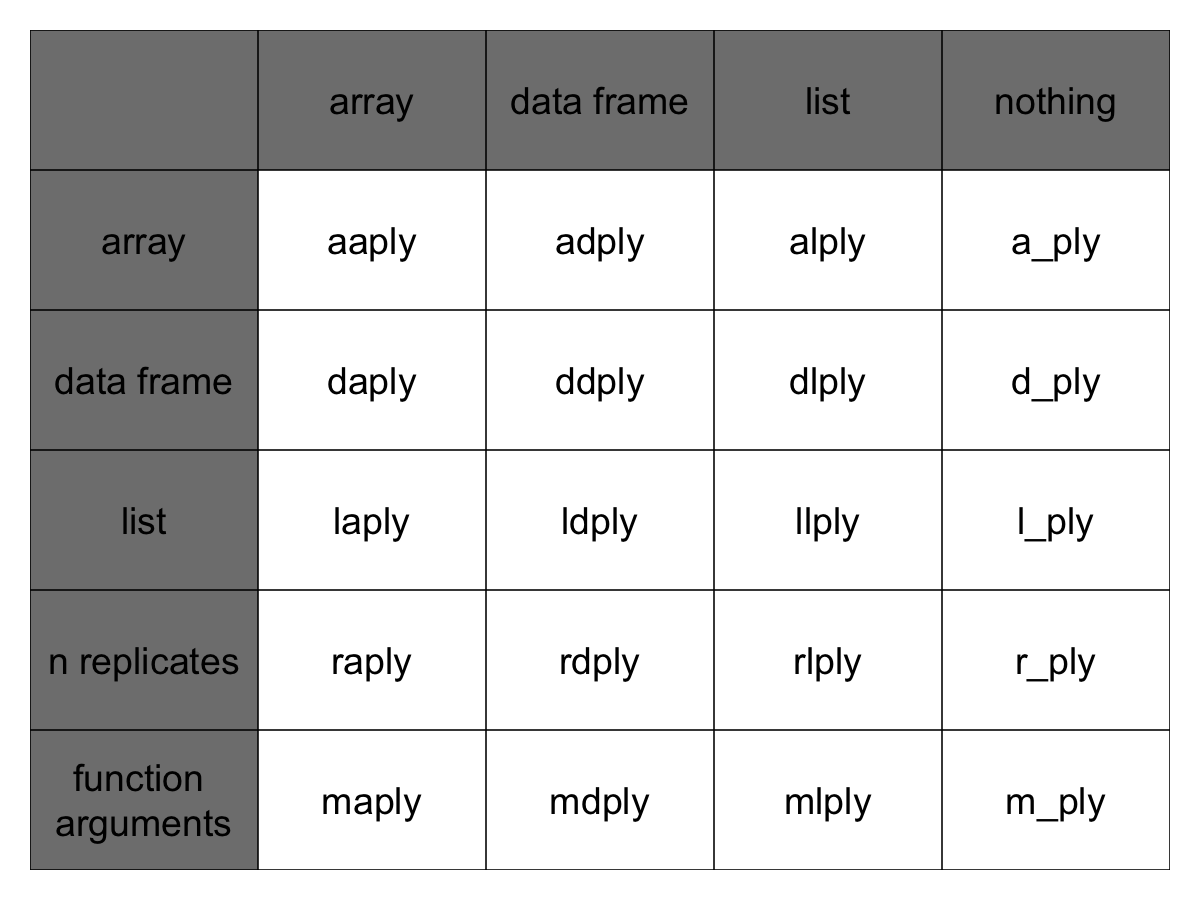
Full apply suite
Each of the xxply functions (daply, ddply, llply, laply, …) has the same structure and has 4 key features and structure:
xxply(.data, .variables, .fun)- The first letter of the function name gives the input type and the second gives the output type.
- .data - gives the data object to be processed
- .variables - identifies the splitting variables
- .fun - gives the function to be called on each piece
Now we can quickly calculate the mean GDP per continent:
ddply(
.data = calcGDP(gapminder),
.variables = "continent",
.fun = function(x) mean(x$gdp)
) continent V1
1 Africa 20904782844
2 Americas 379262350210
3 Asia 227233738153
4 Europe 269442085301
5 Oceania 188187105354
Let’s walk through what just happened:
- The
ddplyfunction feeds in adata.frame(function starts with d) and returns anotherdata.frame(2nd letter is a d) i - the first argument we gave was the data.frame we wanted to operate on: in this case the gapminder data. We called
calcGDPon it first so that it would have the additionalgdpcolumn added to it. - The second argument indicated our split criteria: in this case the “continent” column. Note that we just gave the name of the column, not the actual column itself like we’ve done previously with subsetting. Plyr takes care of these implementation details for you.
- The third argument is the function we want to apply to each grouping of the data. We had to define our own short function here: each subset of the data gets stored in
x, the first argument of our function. This is an anonymous function: we haven’t defined it elsewhere, and it has no name. It only exists in the scope of our call toddply.
What if we want a different type of output data structure?:
dlply(
.data = calcGDP(gapminder),
.variables = "continent",
.fun = function(x) mean(x$gdp)
)$Africa
[1] 20904782844
$Americas
[1] 379262350210
$Asia
[1] 227233738153
$Europe
[1] 269442085301
$Oceania
[1] 188187105354
attr(,"split_type")
[1] "data.frame"
attr(,"split_labels")
continent
1 Africa
2 Americas
3 Asia
4 Europe
5 Oceania
We called the same function again, but changed the second letter to an l, so the output was returned as a list.
We can specify multiple columns to group by:
ddply(
.data = calcGDP(gapminder),
.variables = c("continent", "year"),
.fun = function(x) mean(x$gdp)
) continent year V1
1 Africa 1952 5992294608
2 Africa 1957 7359188796
3 Africa 1962 8784876958
4 Africa 1967 11443994101
5 Africa 1972 15072241974
6 Africa 1977 18694898732
7 Africa 1982 22040401045
8 Africa 1987 24107264108
9 Africa 1992 26256977719
10 Africa 1997 30023173824
11 Africa 2002 35303511424
12 Africa 2007 45778570846
13 Americas 1952 117738997171
14 Americas 1957 140817061264
15 Americas 1962 169153069442
16 Americas 1967 217867530844
17 Americas 1972 268159178814
18 Americas 1977 324085389022
19 Americas 1982 363314008350
20 Americas 1987 439447790357
21 Americas 1992 489899820623
22 Americas 1997 582693307146
23 Americas 2002 661248623419
24 Americas 2007 776723426068
25 Asia 1952 34095762661
26 Asia 1957 47267432088
27 Asia 1962 60136869012
28 Asia 1967 84648519224
29 Asia 1972 124385747313
30 Asia 1977 159802590186
31 Asia 1982 194429049919
32 Asia 1987 241784763369
33 Asia 1992 307100497486
34 Asia 1997 387597655323
35 Asia 2002 458042336179
36 Asia 2007 627513635079
37 Europe 1952 84971341466
38 Europe 1957 109989505140
39 Europe 1962 138984693095
40 Europe 1967 173366641137
41 Europe 1972 218691462733
42 Europe 1977 255367522034
43 Europe 1982 279484077072
44 Europe 1987 316507473546
45 Europe 1992 342703247405
46 Europe 1997 383606933833
47 Europe 2002 436448815097
48 Europe 2007 493183311052
49 Oceania 1952 54157223944
50 Oceania 1957 66826828013
51 Oceania 1962 82336453245
52 Oceania 1967 105958863585
53 Oceania 1972 134112109227
54 Oceania 1977 154707711162
55 Oceania 1982 176177151380
56 Oceania 1987 209451563998
57 Oceania 1992 236319179826
58 Oceania 1997 289304255183
59 Oceania 2002 345236880176
60 Oceania 2007 403657044512
daply(
.data = calcGDP(gapminder),
.variables = c("continent", "year"),
.fun = function(x) mean(x$gdp)
) year
continent 1952 1957 1962 1967
Africa 5992294608 7359188796 8784876958 11443994101
Americas 117738997171 140817061264 169153069442 217867530844
Asia 34095762661 47267432088 60136869012 84648519224
Europe 84971341466 109989505140 138984693095 173366641137
Oceania 54157223944 66826828013 82336453245 105958863585
year
continent 1972 1977 1982 1987
Africa 15072241974 18694898732 22040401045 24107264108
Americas 268159178814 324085389022 363314008350 439447790357
Asia 124385747313 159802590186 194429049919 241784763369
Europe 218691462733 255367522034 279484077072 316507473546
Oceania 134112109227 154707711162 176177151380 209451563998
year
continent 1992 1997 2002 2007
Africa 26256977719 30023173824 35303511424 45778570846
Americas 489899820623 582693307146 661248623419 776723426068
Asia 307100497486 387597655323 458042336179 627513635079
Europe 342703247405 383606933833 436448815097 493183311052
Oceania 236319179826 289304255183 345236880176 403657044512
You can use these functions in place of for loops (and its usually faster to do so): just write the body of the for loop in the anonymous function:
d_ply(
.data=gapminder,
.variables = "continent",
.fun = function(x) {
meanGDPperCap <- mean(x$gdpPercap)
print(paste(
"The mean GDP per capita for", unique(x$continent),
"is", format(meanGDPperCap, big.mark=",")
))
}
)[1] "The mean GDP per capita for Africa is 2,193.755"
[1] "The mean GDP per capita for Americas is 7,136.11"
[1] "The mean GDP per capita for Asia is 7,902.15"
[1] "The mean GDP per capita for Europe is 14,469.48"
[1] "The mean GDP per capita for Oceania is 18,621.61"
Challenge 1
Calculate the average life expectancy per continent. Which has the longest? Which had the shortest?
Challenge 2
Calculate the average life expectancy per continent and year. Which had the longest and shortest in 2007? Which had the greatest change in between 1952 and 2007?
Advanced Challenge
Calculate the difference in mean life expectancy between the years 1952 and 2007 from the output of challenge 2 using one of the plyr functions.
Alternate Challenge if class seems lost
Without running them, which of the following will calculate the average life expectancy per continent:
ddply(
.data = gapminder,
.variables = gapminder$continent,
.fun = function(dataGroup) {
mean(dataGroup$lifeExp)
}
)ddply(
.data = gapminder,
.variables = "continent",
.fun = mean(dataGroup$lifeExp)
)ddply(
.data = gapminder,
.variables = "continent",
.fun = function(dataGroup) {
mean(dataGroup$lifeExp)
}
)adply(
.data = gapminder,
.variables = "continent",
.fun = function(dataGroup) {
mean(dataGroup$lifeExp)
}
)